Query a list of genes in LINCS dataset
"Genes" pipeline allows a query of genes of interest against LINCS data. One may start a query by inputing a list of genes via Entrez gene IDs or gene symbols separated by comma in the search field and clicking "Search for set of gene symbols or IDs". This section describes how to submit your own list of genes for analysis.
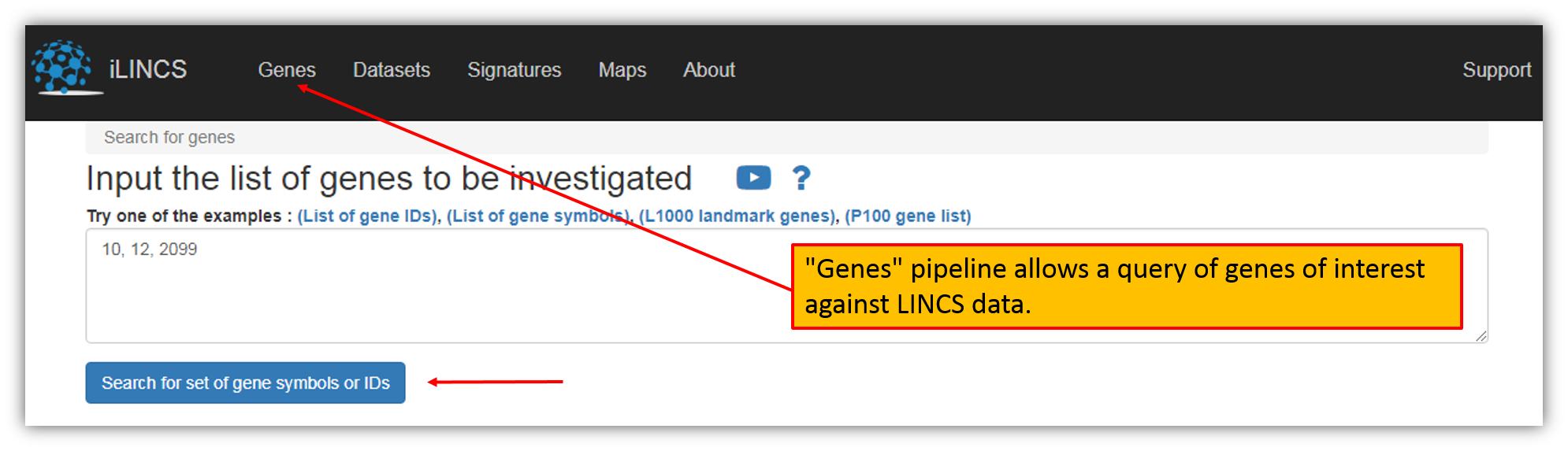
The figure above shows the screen to submit your own list of genes. You can use either entrez ids (e.g. 2099) or symbols (e.g. esr1). As shown in the figure, type/paste a list of genes in the box. Let's input genes 10,12,2099 in the search box as an example and click "Search for set of gene symbols or IDs".
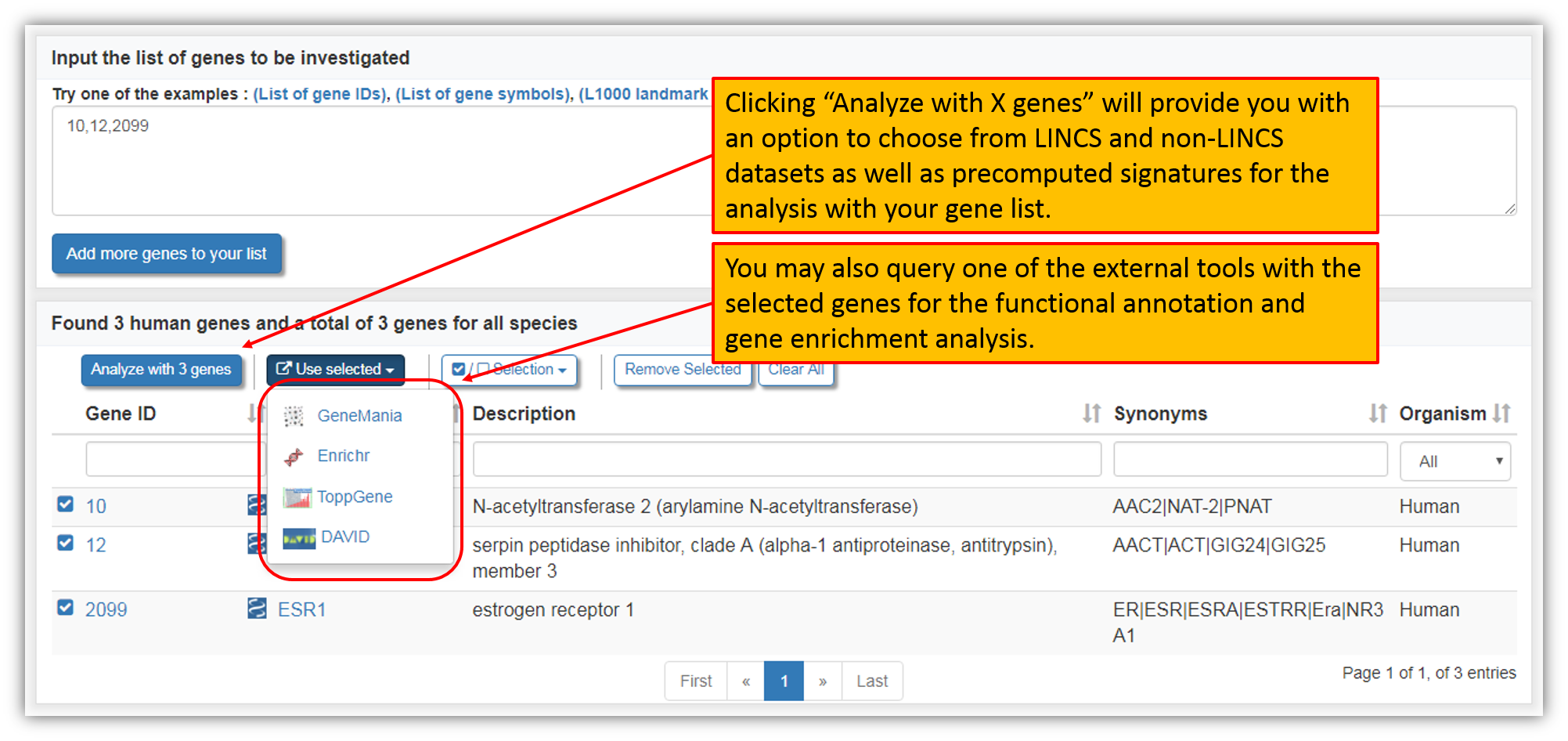
At this point our database is searched for all the genes submitted and the figure above shows the list of gene matches found. To analyze gene(s), select genes of interest (default option is preselected for all found human genes) and then click "Analyze with X genes". You may also submit a query to one of the external tools (Genemania, EnrichR, ToppGene, DAVID) with your gene list at this point.
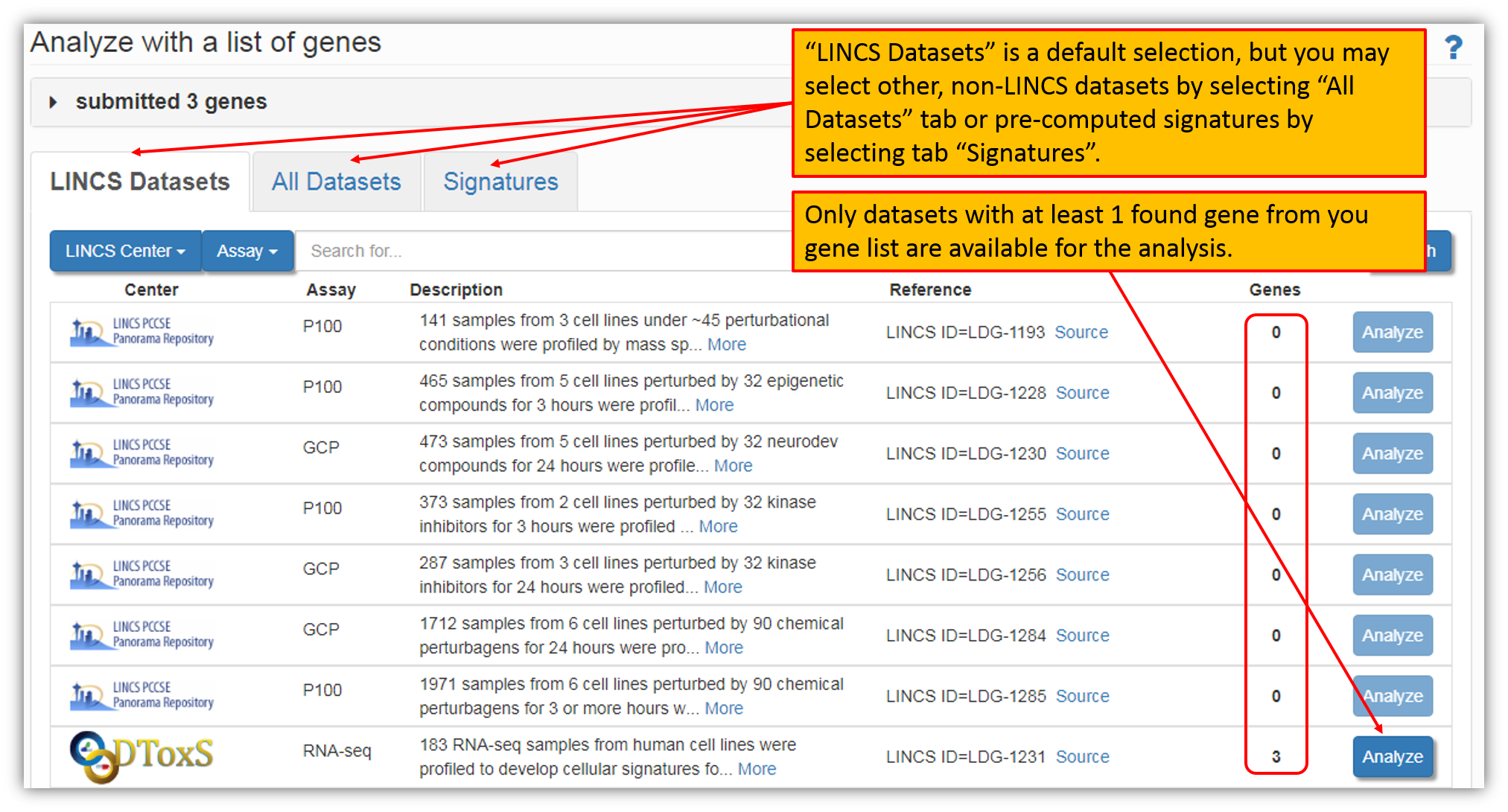
Clicking "Analyze with X genes" button (figure above) and as shown in the figure below, will provide you with an option to choose from LINCS and non-LINCS datasets as well as precomputed signatures for the analysis with your gene list. Default selection is listing all LINCS datasets, but you may explore other datasets by selecting tab "All Datasets" and explore available precomputed signatures by selecting the tab "Signatures". "Genes" column in the figure below shows the number of genes from your query that was found within a particular dataset. Only datasets with at least one found gene will be available for the analysis. Select a LINCS dataset of interest and click "Analyze" next to a it to perform gene list analysis within a dataset.
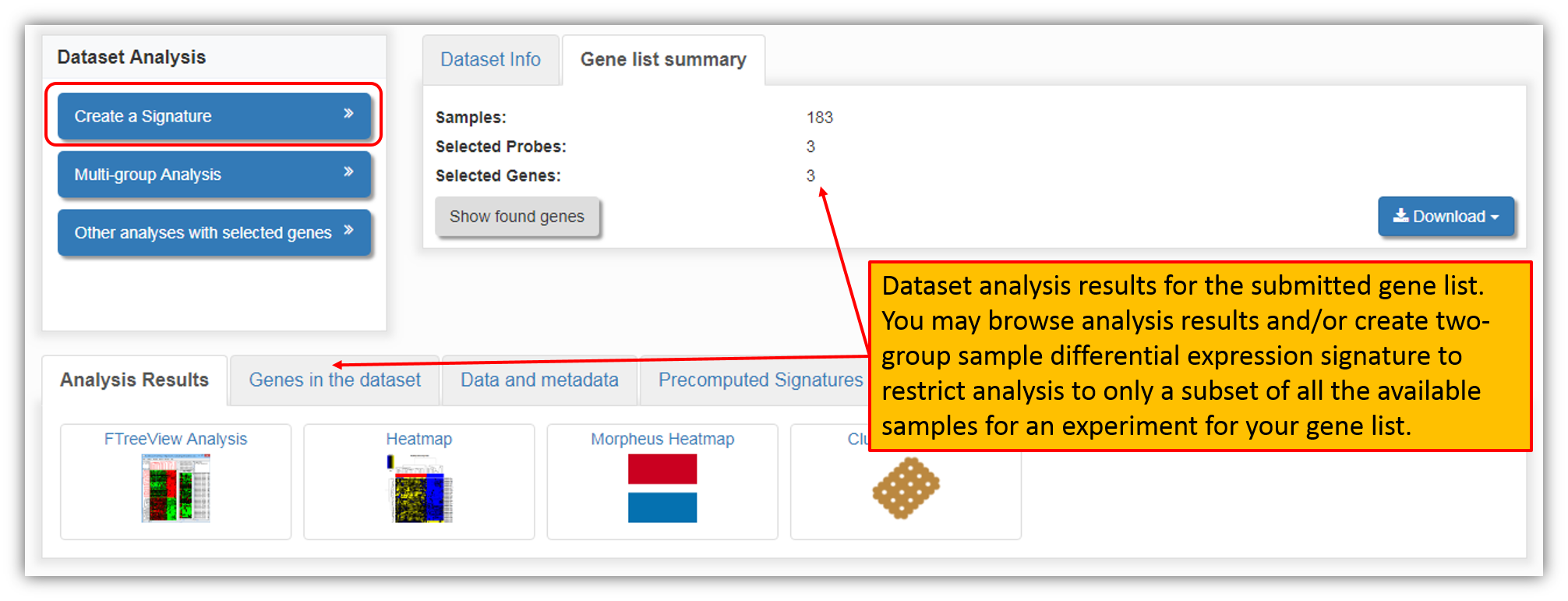
Clicking "Analyze" next to the dataset of interest, will perform a query of the gene list within the dataset and will display analysis results as seen in the figure below. You may also wish to restrict gene list analysis to only a subset of all the available samples for an experiment within a dataset. To do this, you may create a two-group sample differential expression signature by clicking "Create Signature".
 GR calculator
GR calculator GREIN
GREIN Signeta
Signeta