Analyze LINCS transcriptomic and proteomic datasets
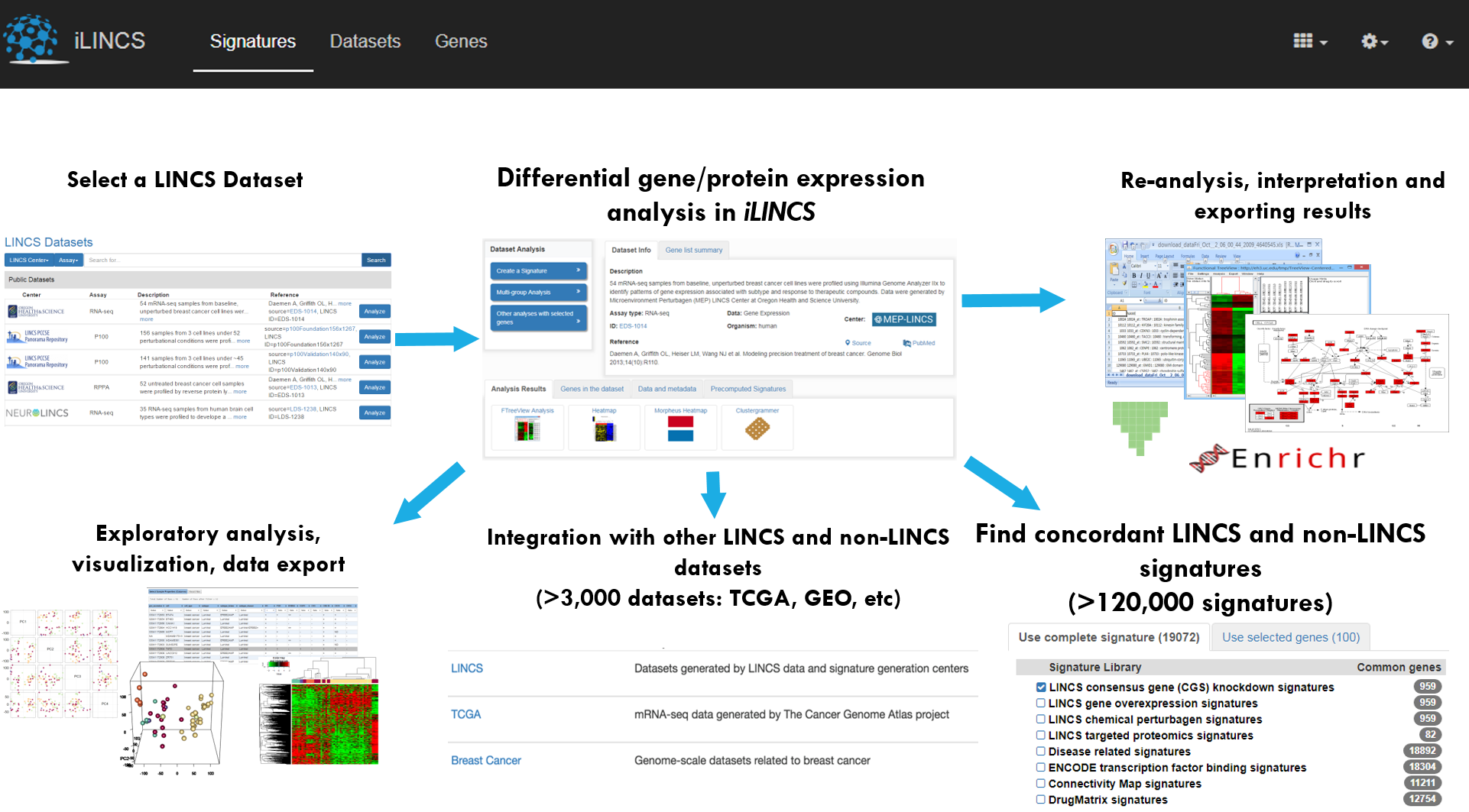
The iLINCS (Integrative LINCS) portal is an web platform for analysis of LINCS data and signatures. The portal provides biologists-friendly user interfaces for analyzing transcriptomics and proteomics LINCS datasets. iLINCS web tools facilitate statistical analysis to identify differentially expressed genes and proteins; bioinformatics analysis to identify affected networks, pathway and gene lists; and interactive visualizations of the results to aid in interpreting your results. The workflow is as follows:
- Select a LINCS dataset (transcriptomic, proteomic, phosphoproteomic)
- Browse pre-computed exploratory analysis results
- Perform statistical analysis to construct a signature of differentially expressed genes or proteins by comparing two groups of samples
- Perform bioinformatics analysis of a created signature
- Interactive visualization of the signature and data used in constructing the signatures
- Find concordant drug, diesese and transcription factor binding signatures (> 120,000 signatures available)
- Perform SPIA pathway analysis
- Perform gene list enrichment analysis
- Use the created signature to query and analyze other LINCS or non-LINCS datasets (> 3,000 datasets available for analysis)
 GR calculator
GR calculator GREIN
GREIN Signeta
Signeta