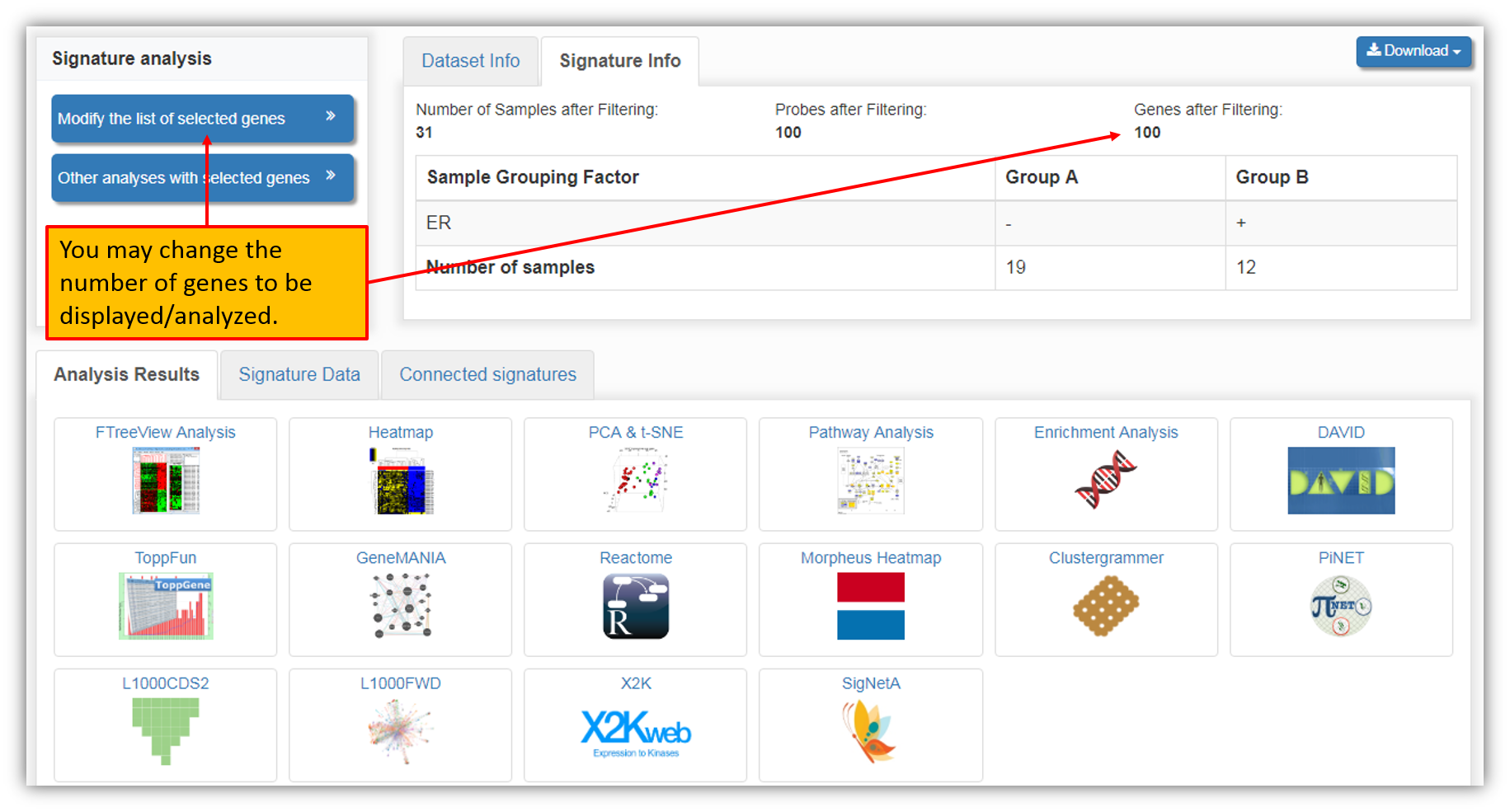
Adjusting number of genes in a signature
"Create a signature" function generates a two-group sample differential expression signature within a selected dataset using top 100 differentially expressed genes between the groups as seen in the figure below. You may adjust the number of included genes for the signature creation. You can select a different set of differentially expressed genes based on different cutoff (fold change and p-values) criteria. This can be accomplished by clicking "Modify the list of selected genes" as seen in the figure below.
Clicking "Modify the list of selected genes" link will open up a Shiny Volcano plot that will allow you to adjust fold change as well as p-values (cutoff values) for the genes to be included in the signature as seen in the figure below.
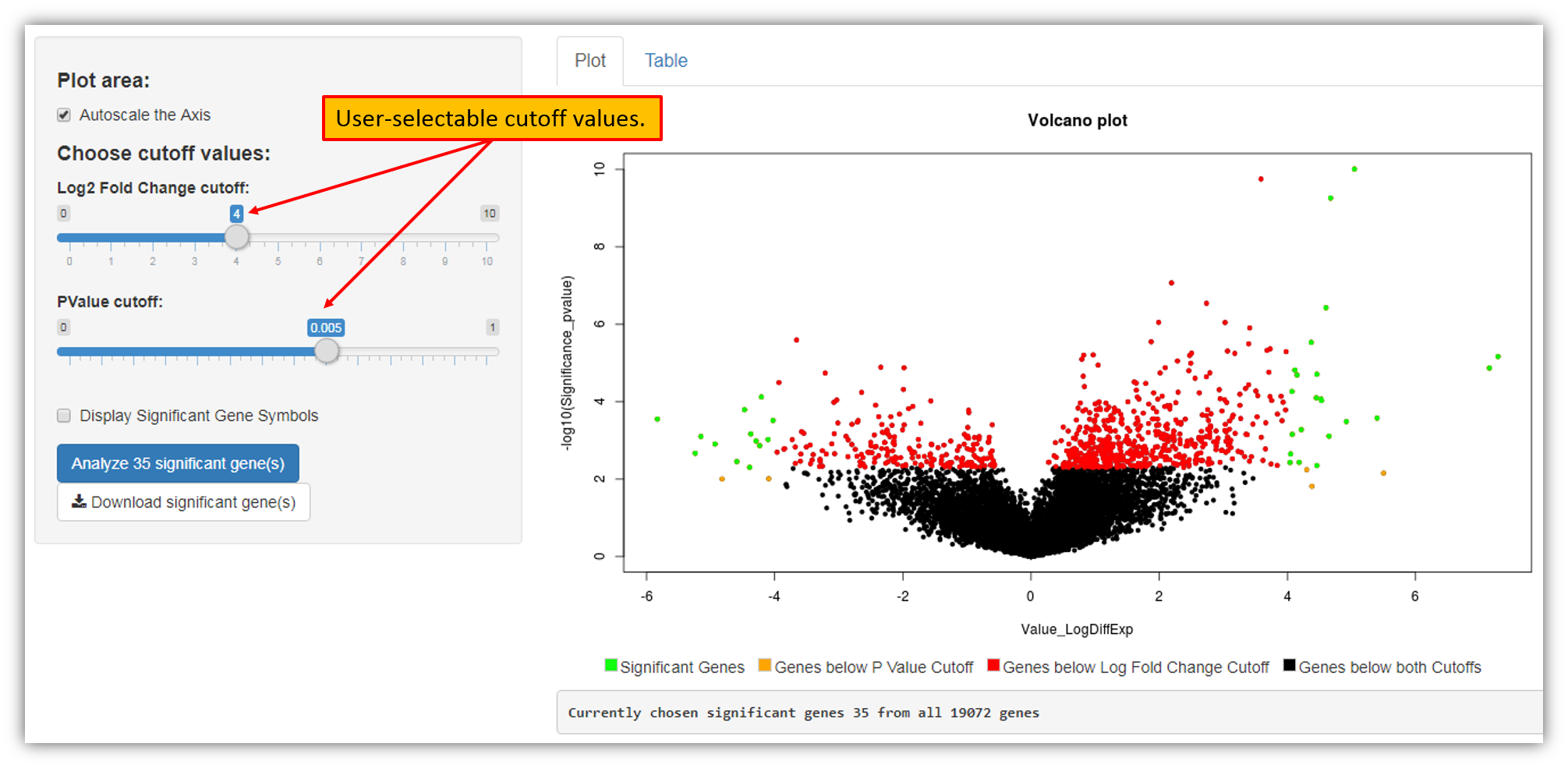
The figure above shows that upon adjusting cutoff values we have decreased the number of genes to be included in the signature from 100 genes (default option) to 35 genes. Clicking "Analyze 35 significant gene(s)" button will re-create a signature using only genes selected using new cutoff criteria.
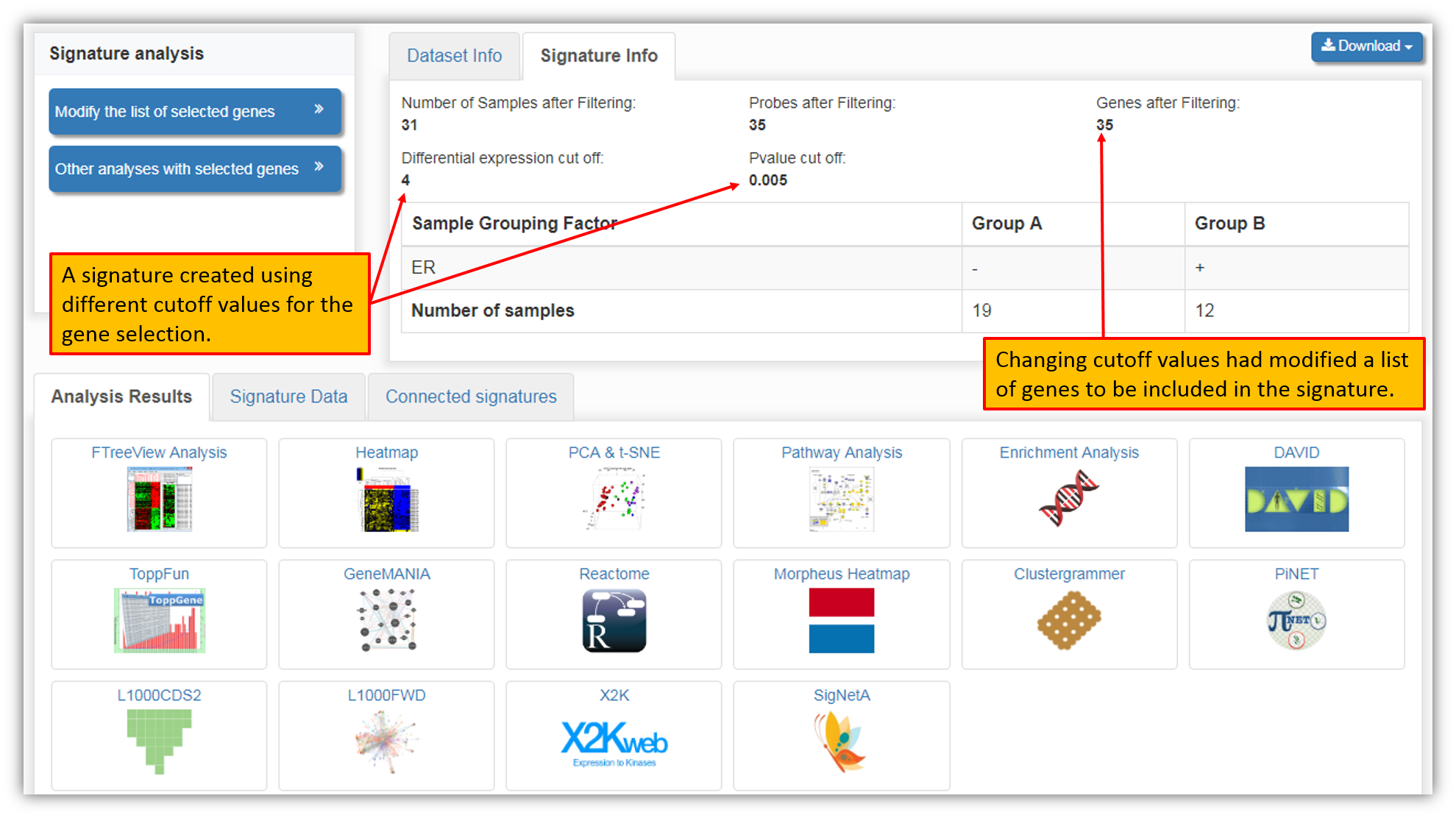
As you can see in the figure above, the signature was re-created using new cutoff criteria for the gene inclusion within a dataset. Just as before, "Analysis Results" were instantly computed for the selected genes within a dataset and are available for viewing and download.
Keywords: tasks,signature |Category: tasks
 GR calculator
GR calculator GREIN
GREIN Signeta
Signeta