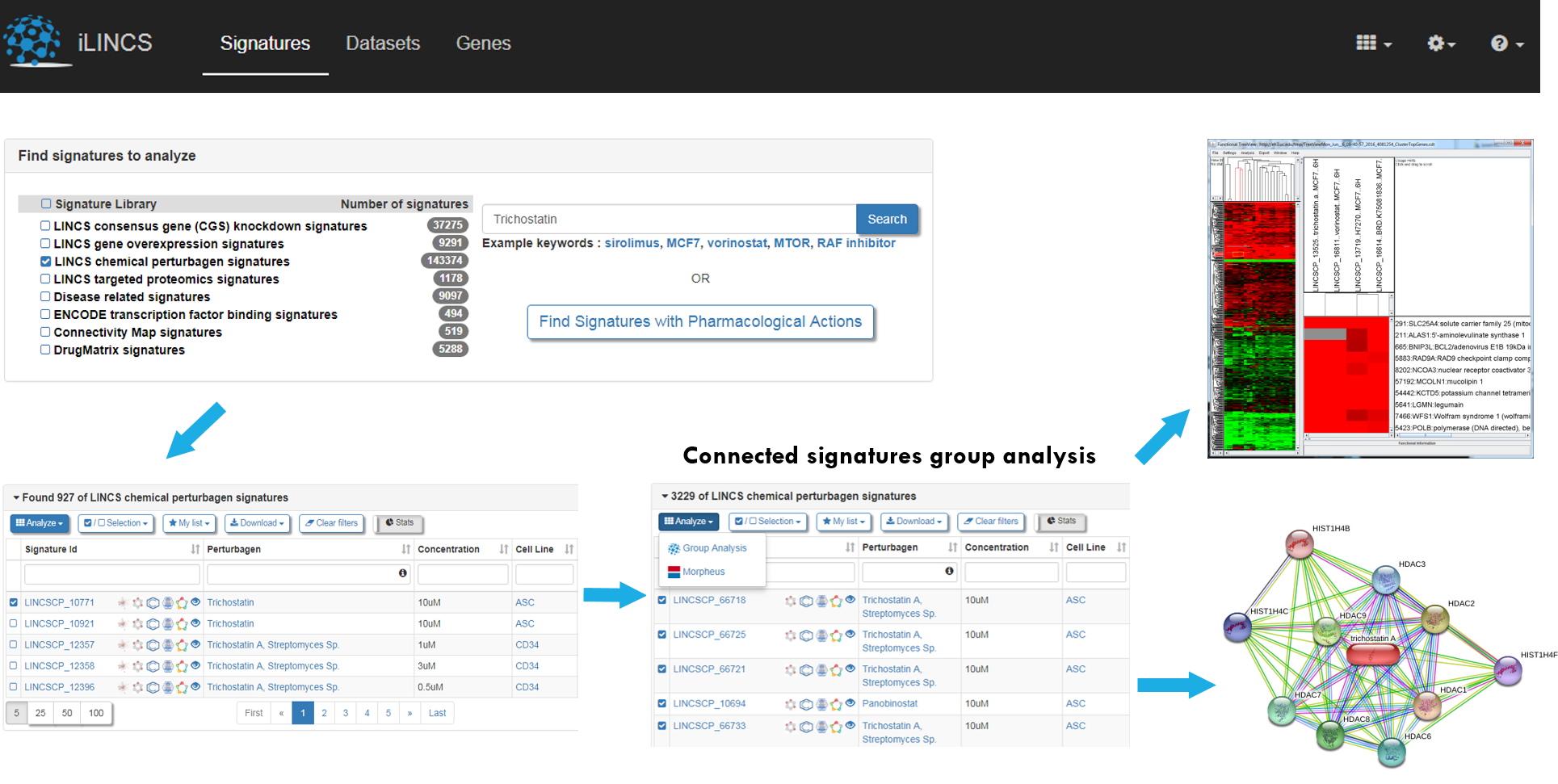
Analyze a drug signature and find other drugs with similar signatures
The iLINCS (Integrative LINCS) portal portal facilitates analysis of transcriptional drug signatures, and search for and analysis of groups of concordant transcriptional signatures of different drugs. The transcriptional signatures of chemical perturbagen activity in the iLINCS portal are constructed based on the Broad L1000 assay data. Each signature consists of the average z-scores and associated p-values for each of 978 landmark genes in the L1000 assay. The user may query perturbagen signatures using a drug name, download a signature of interest, and analyze genes that are up- or down-regulated in response to the drug. A biologist may also perform a comparative analysis with the signatures of other drugs, gene knockdown signatures, and disease-related signatures and try to establish a biological model of signature similarity (concordance). The workflow is as follows:
- Search for a chemical perturbagen or drug of interest, or upload a user-defined signature
- Find signatures concordant with your query signature
- Analyze a group of concordant signatures to study patterns of expression and the biological underpinnings of the observed concordances
- Download a drug signatures for future analysis
 GR calculator
GR calculator GREIN
GREIN Signeta
Signeta