Overview of precomputed signature libraries
In the "Signatures" pipeline, you may explore, analyze and visualize over 200,000 pre-computed signatures (i.e. list of "scores" (activity levels) for a list of genes or for all genes in the genome "genome-wide signatures"). One would land on the Signatures landing page by clicking "Signatures" on the iLINCS portal header.
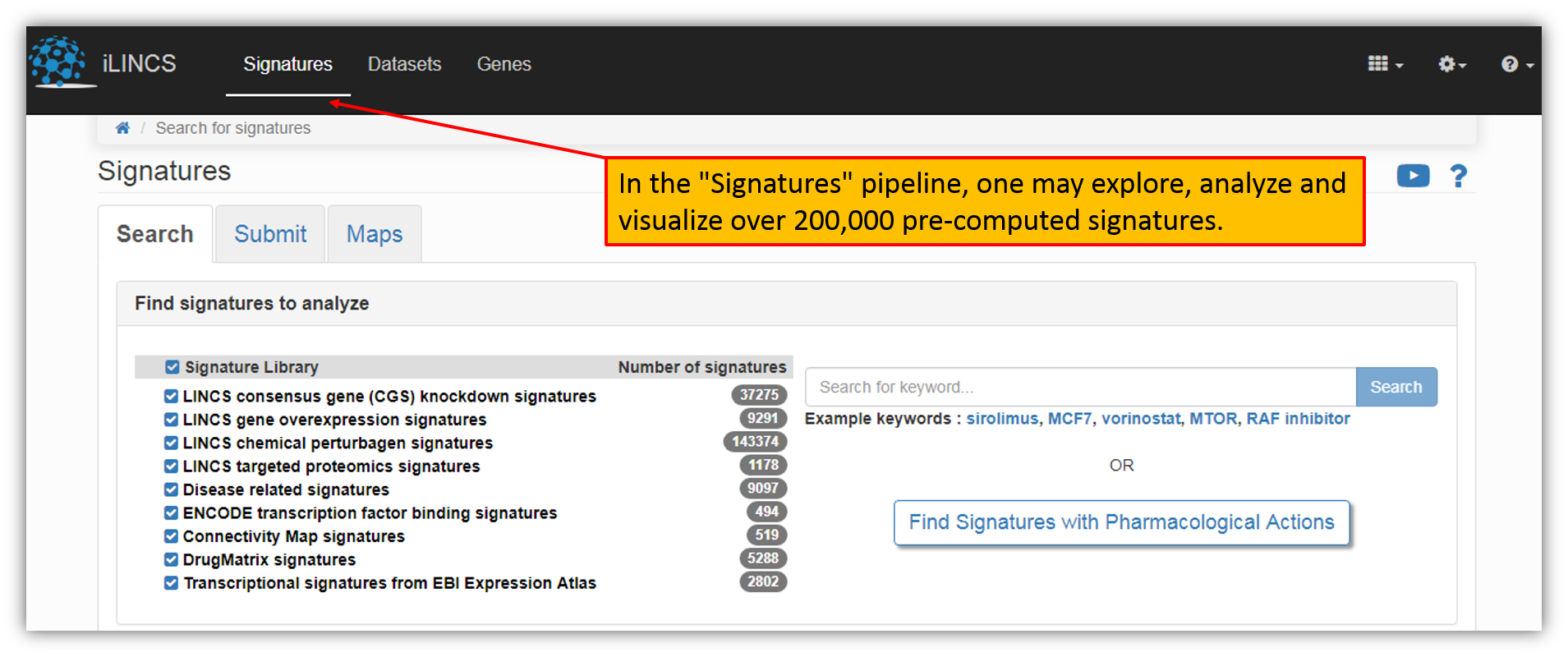
As shown in the figure above, there are 9 pre-computed signature libraries on iLINCS portal.
LINCS consensus gene (CGS) knockdown signatures - Transcriptional signatures were constructed by further aggregating signatures of individual shRNA perturbations. The signatures consist of differential gene expressions and p-values for 978 Landmark Genes measured by L1000 assay.
LINCS gene overexpression signatures - Transcriptional signatures of gene overexpression based on L1000 assay. The signatures consist of differential gene expressions and p-values for 978 Landmark Genes measured by L1000 assay. The signatures were created by aggregating (ie averaging) Level 4 data for biological replicates as defined by the signatures metadata. Only signatures designated to be reproducible and self-connected ("gold") by the Broad institute are represented.
LINCS chemical perturbagen signatures - Transcriptional signatures of perturbations by small molecules based on L1000 assay. Signatures were created by aggregating (ie averaging) Level 4 data for biological replicates as defined by the signatures metadata. Only signatures designated to be reproducible and self-connected ("gold") by the Broad institute are represented. The signatures consist of differential gene expressions and p-values for 978 Landmark Genes measured by L1000 assay.
LINCS targeted proteomics signatures - Signatures of perturbations assayed by P100 against 96 phosphopeptide probes and GCP assay against ~60 probes that monitor combinations of post-translational modifications on histones. The data is generated by using mass spectrometry techniques to characterize proteome level molecular signatures of responses to small molecule and genetic pertubations in a number of different cell lines.
Disease related signatures - Transcriptional signatures constructed by comparing sample groups within the collection of public domain transcriptional dataset (GEO GDS collection). Each signature consists of differential expressions and associated p-values for all genes.
ENCODE transcription factor binding signatures - Transcription factor (TF) binding signatures constructed using ENCODE ChiP-seq data. Each signature consists of genome scale (i.e. for each gene) scores and probabilities of regulation by the given TF in the specific context (cell line and treatment). These signatures were developed using our in-house TREG methodology (paper under review).
Connectivity Map Signatures - Transcriptional signatures of perturbagen activity constructed based on the version 2 of the original Connectivity Map dataset using Affymetrix expression arrays. Each signature consists of differential expressions and associated p-values for all genes when comparing perturbagen treated cell lines with appropriate controls.
DrugMatrix signatures - Transcriptional signatures of DrugMatrix. Signatures consist of differential gene expression and associated p-values for ~ 13,000 genes.
Transcriptional signatures from EBI Expression Atlas - Transcriptional signatures constructed by comparing sample groups within the collection of public domain transcriptional dataset (EBI Array Express collection). Each signature consists of differential expressions and associated p-values for all genes.
Keywords: tutorial |
 GR calculator
GR calculator GREIN
GREIN Signeta
Signeta